Data Pipeline#
Note
This page documents the pipeline developed within the scope of the initial MVP.
Executive Summary#
Tip
A data pipeline is an automated system that takes raw data, transforms it in some way, then moves it to a destination storage area.
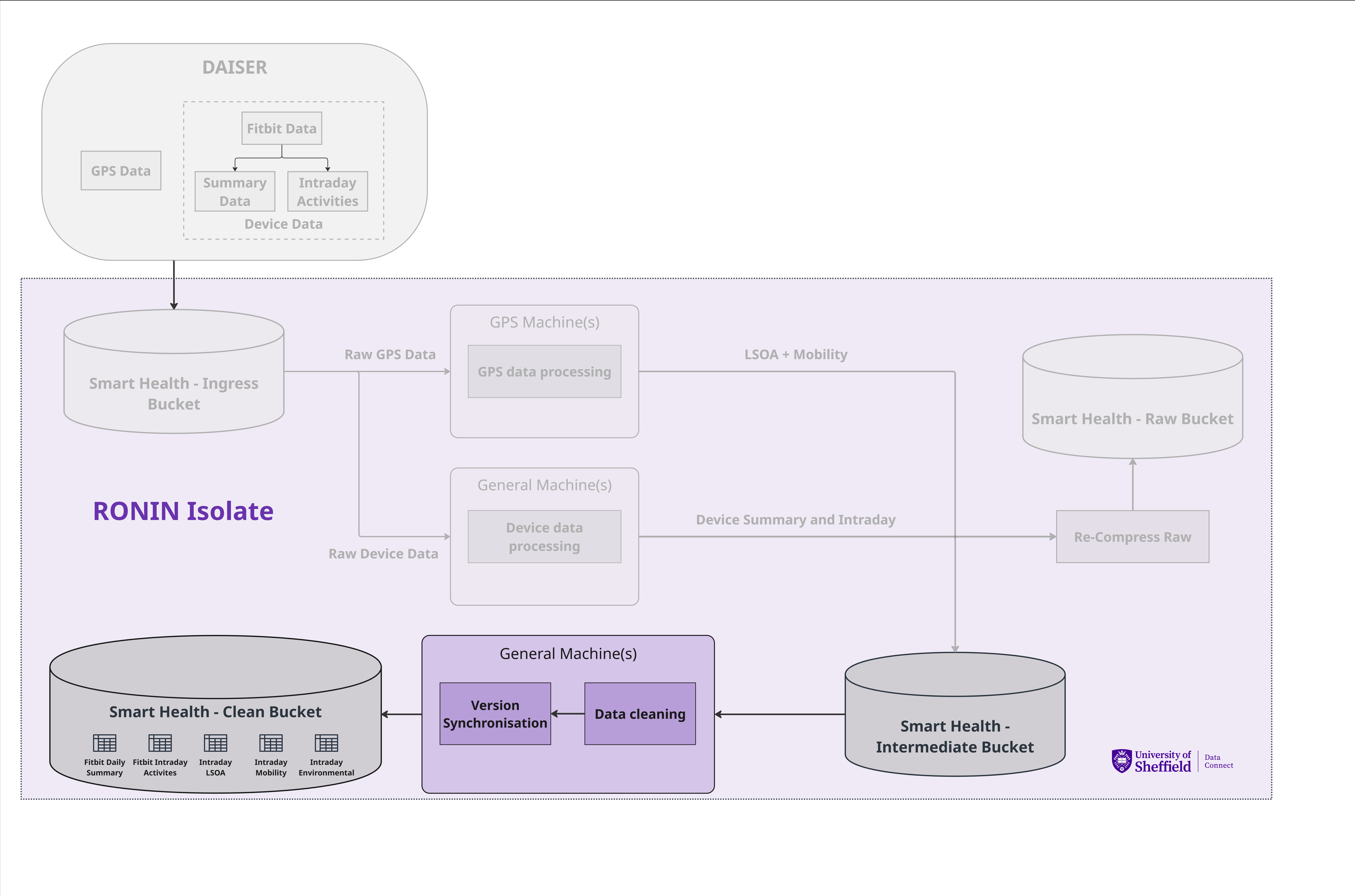
The purpose of the Smart Health MVP data pipeline is to ingest a daily, compressed archive of
.jsondata from Daiser, then clean, enrich, and pseudonymise the data. The processed data will form part of a “research-ready” database in S3 object storage.
Data will be ingressed to our RONIN Isolate environment on a daily basis from Daiser via Secure File Transfer Protocol (SFTP). This compressed archive will be stored in an S3 “Ingress” bucket. Each compressed archive will contain the previous 7 days’ participant data for both GPS data and Fitbit data. The Fitbit data will be comprised of daily summary data, and intraday (many samples within one day) activity data.
For data governance purposes, the highly-sensitive GPS data will be processed on a separate machine to other data. In this stage, after data validation checks, the GPS data will be converted to LSOA, and mobility measures derived. The Fitbit Device data is validated and processed on another machine. The processed data is saved to an S3 “Intermediate” bucket.
The raw data is re-compressed and saved to a “Raw” bucket.
These steps comprise Stage 1: Ingress to Intermediate.
In Stage 2: Intermediate to Clean, the pseudonymised LSOA data can be processed alongside other data steams. In this stage, the LSOA data is enriched with AHAH data. All processed data streams are then merged into the clean database, stored in the “Clean” S3 bucket.
This process is outlined in the diagram below.
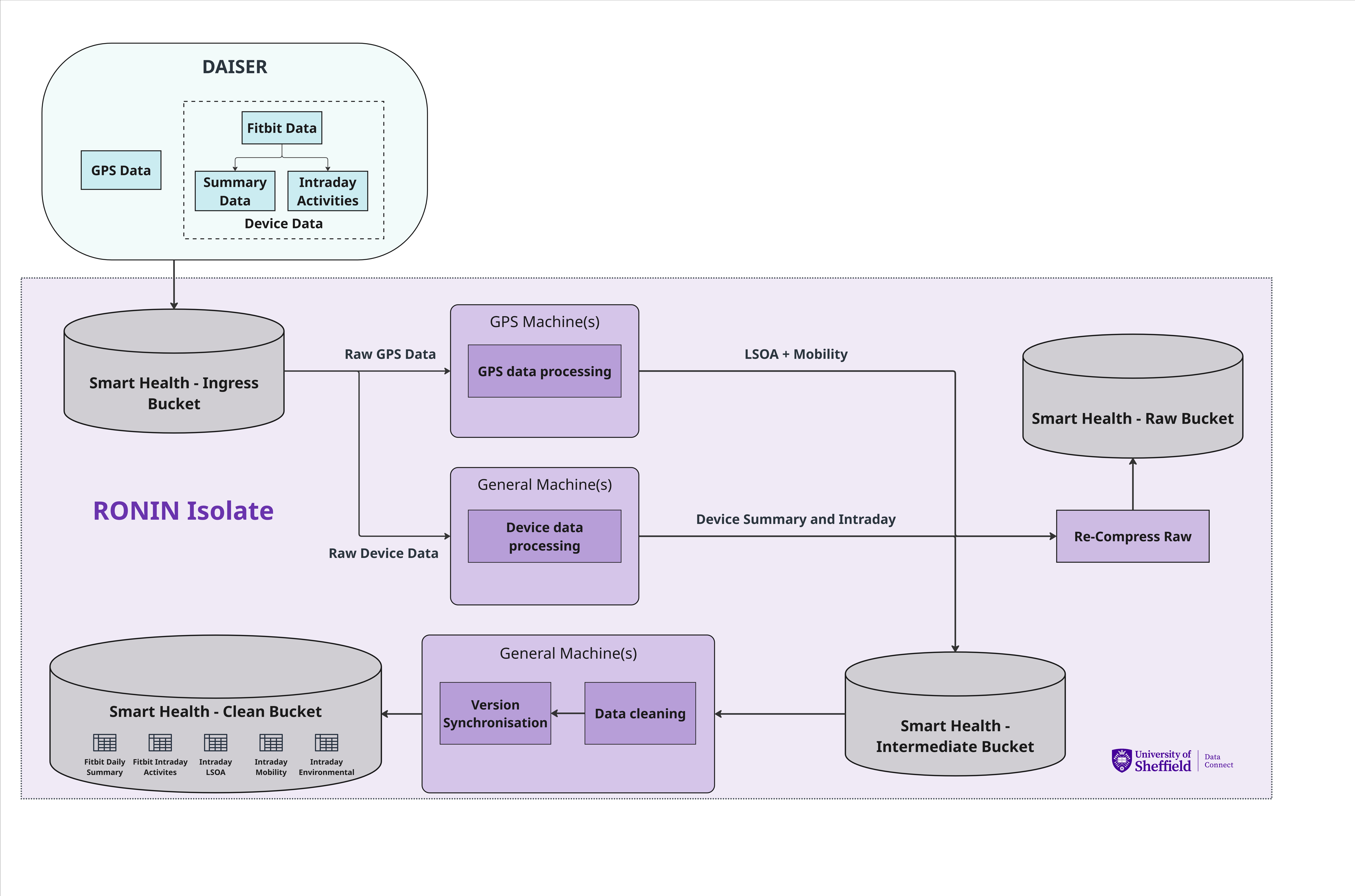
A diagram of the Smart Health MVP pipeline.#
Technical Specification#
The Smart Health pipeline is a multi-stage, batch, ETL pipeline with medallion lakehouse architecture. As data moves through the pipeline its quality is progressivley improved, from the “Raw” bucket (Bronze layer), to the validated, pseudonymised “Intermediate” bucket (Silver layer), through to the enriched, research-ready “Clean” bucket (Gold layer).
Data Source#
Ingestion method: SFTP into S3 object storage bucket in RONIN Isolate
Format (variety): .json within compressed .7z archive
Frequency (velocity): Daily. Each compressed .7z archive will contain the previous 7-days of data.
Volume: up to 10,000 participants, one .json file per participant, per day, per stream i.e. GPS, Fitbit Daily Summary, Fitbit Intraday activities.
Data Loading Strategy: Currently Full Load with a Change Data Capture (CDC) mechanism. The full archive of 7 previous days is processed and merged into the database. In future stages, this will move to an Incremental (delta) Load, where only data within the archive that has actually been changed will be synchronised. More details can be found in Data Versioning.
Infrastructure and Configuration#
Processing framework: Batch processing using Python scripts managed by a workflow manager. All scripts are available under an MIT licence in the source repository.
Environment: RONIN Isolate AWS Secure Data Environment
Dependency Management: All Python dependencies are managed using the open-source UV package manager1UV is an extremely fast Python package and project manager written in Rust. See here for more information. for fast, reliable, and reproducible virtual environment builds.
Resource requirements: Tested using EC2 VMs with 32Gb RAM, 4 vCPUs, and 30Gb SSD operating on Ubuntu 22.04
Configuration: Database and Workflow manager configured seperately, see Customising the Pipeline.
Transformation Logic#
Validation:
Validation of the raw .json data is applied using the pydantic Python library. For each raw data stream, a list of expected data fields and their expected data types are defined. Where possible, pydantic will coerce the data into the specified data type.
Files containing unexpected fields are rejected and skipped for Information Governance purposes.
GPS data:
GPS data files will contain an array of intraday data points. Expects datetime, latitude, and longitude fields for each GPS data point in the following format:
d: datetime
long: float
lat: float
Unfeasible latitude / longitude values (outside of -90 to 90, and -180 to 180, respectively) are filtered from the data at this stage
Fitbit data:
Daily Summary data is expected to contain a number of fields in the following format:
timestamp: date
calories: int
caloriesBMR: int
distance: int
elevation: int
floors: int
minutesSedentary: int
minutesLightlyActive: int
minutesFairlyActive: int
minutesVeryActive: int
steps: int
breathingRate: int
vo2Max: int
dailyRmssd: int
deepRmssd: int
sleep_deep_minutes: int
sleep_light_minutes: int
sleep_rem_minutes: int
sleep_wake_minutes: int
sleep_minutesToFallAsleep: int
sleep_timeInBed: int
SP02_avg: int
SP02_min: int
SP02_max: int
tempCore: int
Intraday Activities data files will contain an array of intraday data points in the following format:
d: datetime
v: int
d is mapped to the field name timestamp. v is mapped to the field name value.
Cleaning:
Unrealistic speed of travel values (derived from GPS data) are filtered. By default this is any speed greater than 36 metres per second.
Where there is a large time jump between GPS data points (by default greater than 5 minutes), the distance travelled and speed of travel will not be calculated.
The LSOA value at these invalid data points is also filtered
Pseudonymisation:
GPS data is highly sensitive information, and will therefore be pseudonymised by conversion to LSOA values.
Aggregation:
Raw intraday data points are aggregated to a repeatable resolution. This resolution must balance data size/ storage considerations with the granularity required for research purposes. By default, intraday measures (LSOA, mobility, Fitbit activites) are aggregated into 5-minute time periods.
The Environmental data is aggregated to 1-hour time periods.
Some Intraday Fitbit activity data is aggregated over a 24 hour period to produce additional daily summary metrics, such as the average (mean) heart rate per day.
The specific (default) aggregate calculations used to produce each table, and any filters applied during calculation, are as follows:
Intraday LSOA:
Aggregated into 5-minute periods. The ‘latest’ LSOA value is taken within each 5-minute period, defined as the most-recent LSOA at a valid data point (i.e. note after a large time jump.)
Intraday Mobility:
Aggregated into 5-minute periods, including:
The average (mean) speed of travel within each time period
The total (sum) distance travelled within each time period
The number (count) of samples in each time period
The number (count) of unique LSOAs in each time period
Intraday Environmental:
Aggregated into 1-hour periods. These aggregates are calculated from the aggregated Intraday LSOA data points. The modal LSOA is calculated for each time period. Where a tie-break exists, the latest (most recent) LSOA will be used.
Fitbit Intraday Activities:
Aggregated into 5-minute periods and 24 hour period:
Heart rate:
Average (mean) heart rate for each 5-minute and 24 hour period
Minimum heart rate for each 5-minute and 24 hour period
Maximum heart rate for each 5-minute and 24 hour period
The number (count) of samples in each 5-minute time period
Steps:
The total number of steps taken for each 5-minute and 24 hour period
The number (count) of samples in each 5-minute time period
Fitbit Daily Summary: Naturally a single, daily metric. No aggregation is applied.
Enrichment:
All tables except Fitbit Daily Summary will have a
date,time, anddatetimefieldAHAH data used to derive Environmental data table from LSOA values
A
SheffieldID, unique to each participant, is attached to each record
Destination#
Target system: AWS S3 data lakehouse in a RONIN Isolate Secure Data Environment
Output tables:
Intraday LSOA
Intraday Environmental
Intraday Mobility
Fitbit Daily Summary
Fitbit Intraday Activities
File Format: Apache Parquet
Update strategy: Upsert/merge
Schema enforcement: Flexible, as parquet file contains its own schema metadata
Partioning: Tables will be partitioned based on time (year, month, and day)
Structure: s3://<clean bucket>/<table name>/<year>/<month>/<day>/file.parquet. See Directory Structure.
Orchestration#
Workflow management: The pipeline’s processing framework is managed and controlled by Snakemake2snakemake.github.io, a specialized workflow management system based on Python. Snakemake defines the pipeline’s entire control flow as a Directed Acyclic Graph (DAG), where individual transformation steps are represented as rules. This ensures that the complex multi-stage process (Ingress –> Intermediate –> Clean) is executed reliably and reproducibly.
Execution of the Snakemake workflows will be automated using cron scheduled jobs from a shell script.
Retry policy: The Snakemake workflow can be configured to retry failing jobs.
Monitoring, Logging, and Testing:#
Error handling:
Where dates specified for processing are not available in the daily, compressed archive, Snakemake will log a warning and continue with the available dates.
Where files contain additional fields to those expected to be ingressed, a file will be rejected on Data Governance grounds.
Files that fail data validation / quality checks will be skipped.
Logging: The Smart Health pipeline extends Snakemake’s native logging to provide detailed and persistent logs for each workflow execution. Logging is managed using Python’s native logging module.
The path for the log files can be configured separately to the path for data outputs. This facilitates the use of storage-optimised, ephemeral drives to fast I/O data operations, whilst ensuring logs persist in the case of machine failure.
Each log filename contains a timestamp to prevent overwriting.
Two types of log are output for each workflow execution:
A global Snakemake workflow log. This file contains the date that the workflow is executed on.
Rule-specific logs for each rule that executes a Python script. The logging level can be configured from the
config.pyfile. This file is named with the date of the processed day within the compressed archive.
Log files are stored under a top-level logs/ directory. Under this directory is a folder labelled with the date of the daily archive being processed, with the name of the workflow appended.
When the same date is processed in multiple daily archives, the logs for each will be in separate subdirectories (labelled after the archive date).
e.g. for the archive dated 2025-11-11, processing day -1 and -2, the logs from the GPS workflow run (with one script running rule, process_GPS) would output the following:
├── logs
│ └── 2025-11-11_GPS
│ ├── 2025-11-09_08:34:20_process_GPS.log
│ ├── 2025-11-10_08:34:20_process_GPS.log
│ └── 2025-12-09_08:34:25_snakemake_log.log
By default, the format of the log files is "%(asctime)s [%(levelname)s] | %(name)s: %(message)s"3Logging documentation. This format outputs the logs as (if the logging level is configured to INFO):
'YYYY-MM-DD HH:MM:SS,ms [INFO] | bhf_smarthealth.<module>.<filename>: <message>'
Unit testing: Each unit function of the pipeline codebase is unit tested using the Python testing framework pytest.
End-to-end testing: The Snakemake-orchestrated pipeline has been end-to-end tested for 7 days of 10,000 participants’ data to mimic the maximum expected daily, compressed archive size.
Security and Compliance#
[ To be completed, ]
Snakemake Pipeline Workflow Implementation#
A Snakemake workflow is defined by specifying rules in a Snakefile. Rules decompose the workflow into small steps (for example, the application of a single tool) by specifying how to create sets of output files from sets of input files. Snakemake automatically determines the dependencies between the rules by matching file names.
By default, the first rule in a Snakefile is used to define build targets. In the Smart Health Snakefiles, this is named rule all. Snakemake automatically builds a Directed Acyclic Graph (DAG) of the series of rules in the Snakefile to determine the execution order that created the build targets.
Each Snakemake rule essentially says “To create [OUTPUTS], I need these [INPUTS], and I will execute [ACTIONS] to do it.” The actions can be command-line code, or custom Python scripts.
This section details the rules / steps that Snakemake manages for both stages of the Smart Health pipeline.
Data Versioning#
The compressed daily archive recieved from Daiser will contain the 7 previous days’ worth of data. The reason for including the previous 7 days is to capture any data that may have been delayed, corrected, or updated since the last data file. This would be as a result of a participant syncing their wearable device beyond the date that it was collected.
This means that a participants’ data for a particular day will be “seen” up to 7 times by the Smart Health pipeline.
There are two main approaches to re-processing this sliding window of data:
1. Full Load (a.k.a. process everything)
This is the current Data Loading strategy implemented.
All records within the archive are processed by the Smart Health pipeline, irrespective of whether the file contains differences to previous versions. Any changes are upserted into the “Clean” tables.
This is simple to implement, but has high resource usage. This has been minimised by processing a file for only 3 timepoints in the 7-day sliding window.
This is demonstrated in the diagram below.
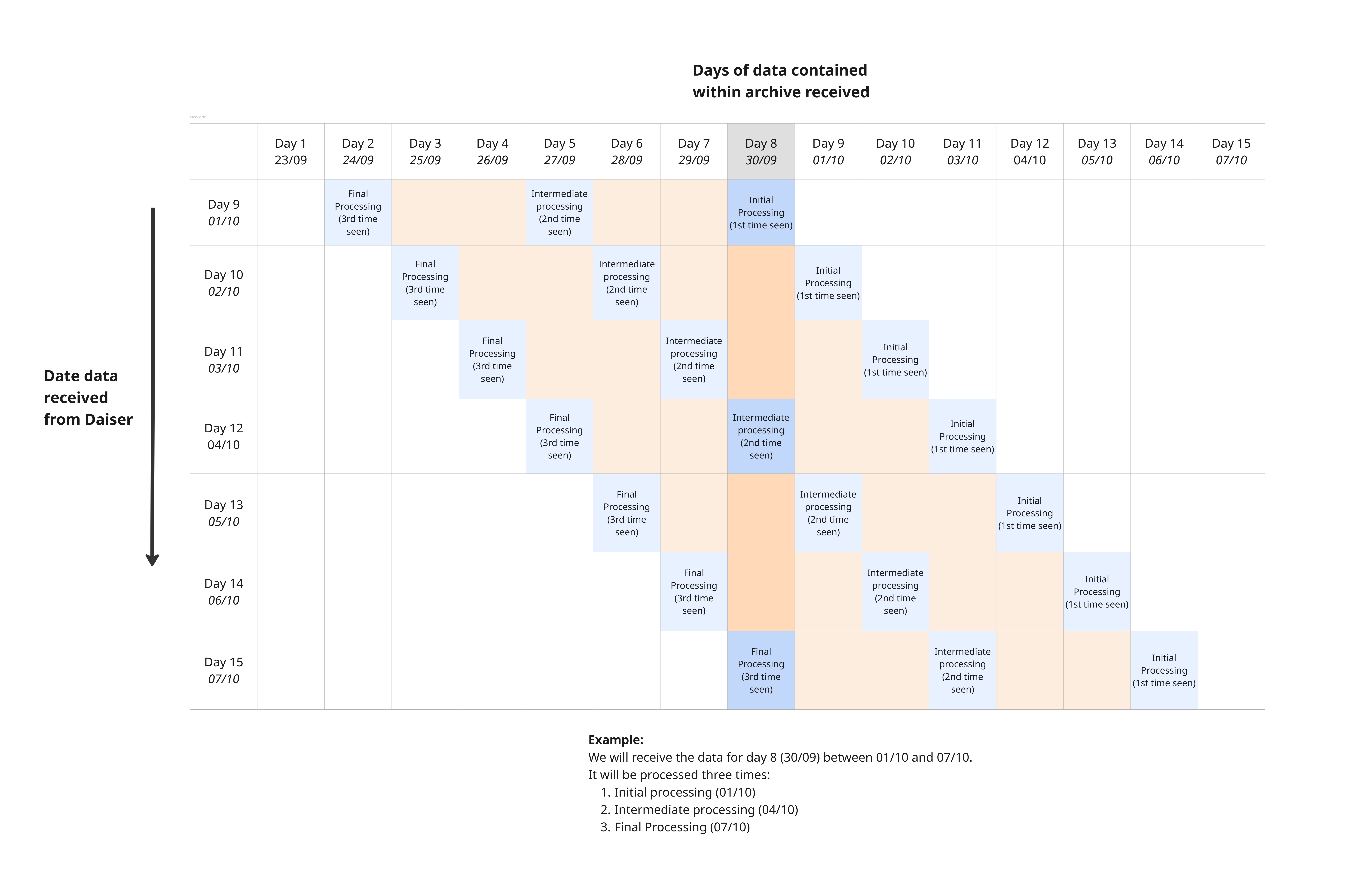
test#
To implement the Full Loading strategy, the workflow/config/config.yaml file contains a XX_DAY_DELTAS key, where XX is the workflow being executed (either GPS or Device). This list of integers indicates which days prior to the receipt date of the archive that should be processed. See Customising the pipeline for more information. This list of deltas is used to dynamically generate the output files expected by the rule all in each Stage 1 workflow.
At this initial workflow stage, a Snakemake checkpoint is run to ensure that the defined deltas are present in the daily archive file. A message is logged to warn if any are not present, and to list the dates that will be processed.
2. Incremental (delta) Load (a.k.a. process new data only)
In a future development phase, the Smart Health will instead implement an incremental load strategy.
The pipeline will implement a filtering strategy that will check each file for changes compared to its previous versions. This will be achieved by comparing the file diff, or by generating a hashed string from each file to check its identity.
Using this approach, only files that have actually changed would be processed by the pipeline, with the changes upserted to the “Clean” tables.
Stage 1: Ingress to Intermediate#
Click here to see the Stage 1 pipeline diagram
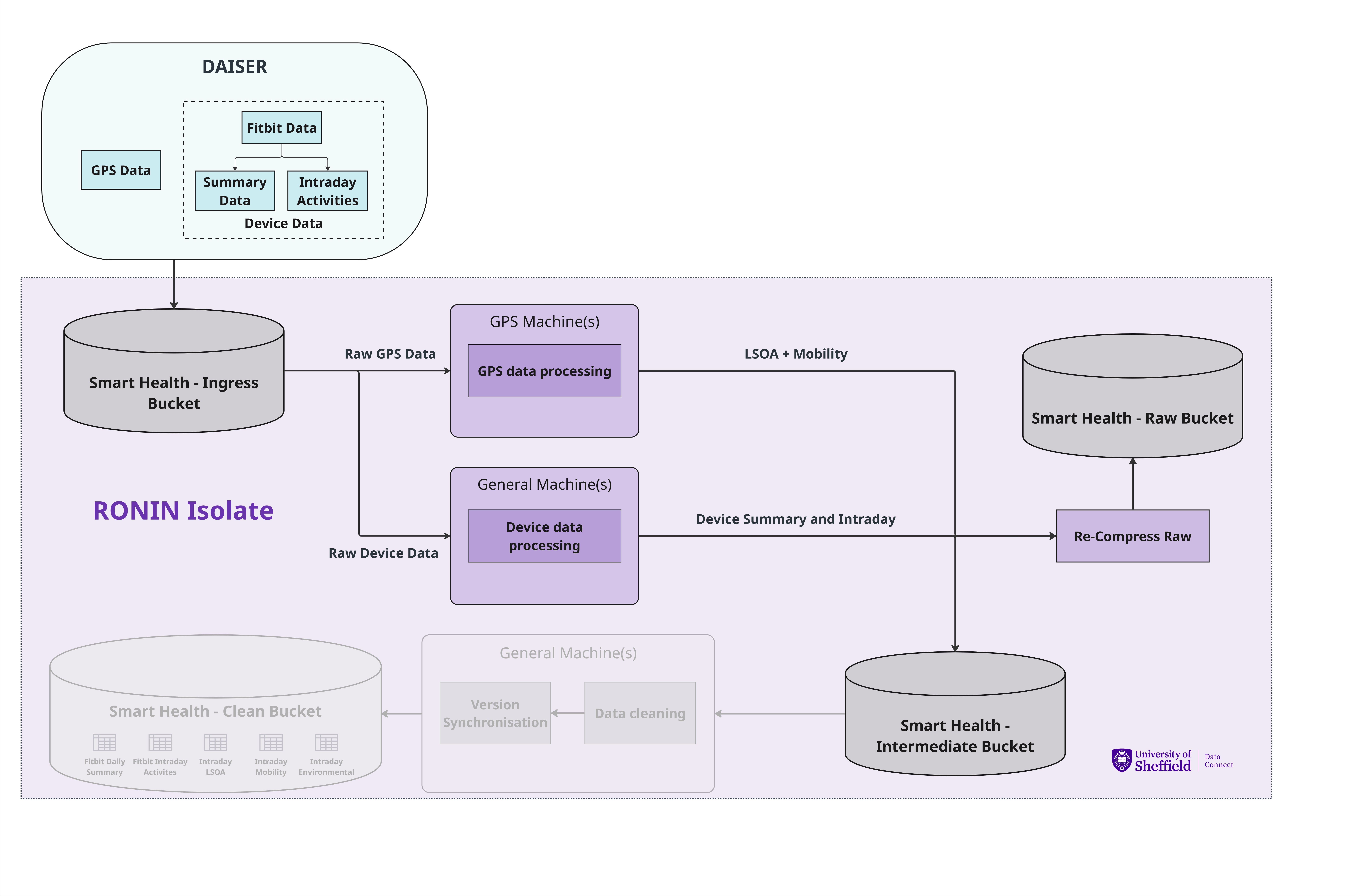
Stage 1 of the Smart Health pipeline#
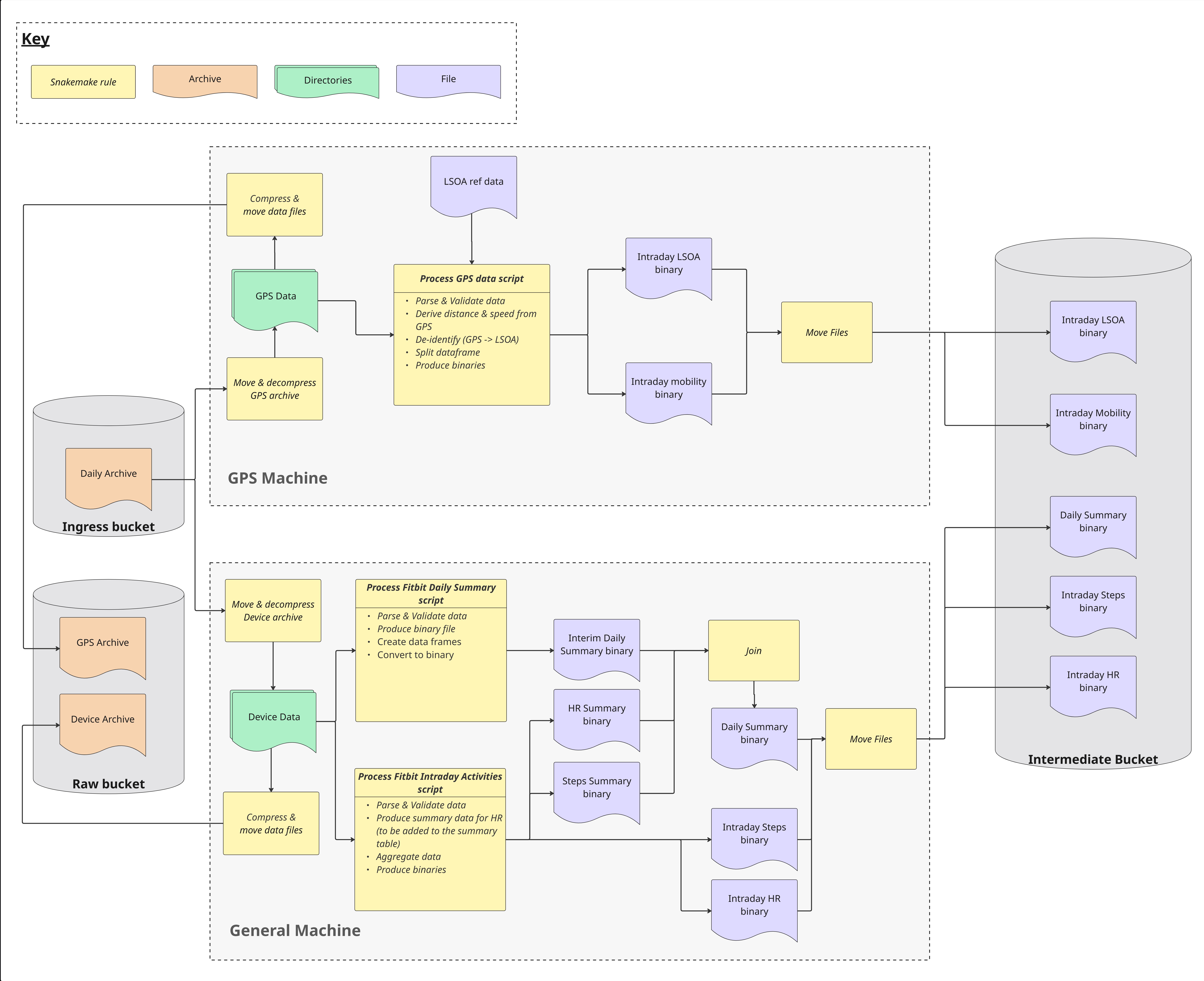
A diagram outlining the Snakemake rules, input and output files, and data flow of Stage 1 of the Smart Health pipeline#
The GPS and Device data processing workflows are seperated into two Snakefiles. This both satisfies Separation of Concerns, and facilities the isolated (and parallel) processing of GPS data on a separate machine.
GPS processing#
The GPS Snakefile runs the following steps:
Download and extract GPS data from the compressed daily archive in the “Ingress” bucket using a
shellcommandReference LSOA data is downloaded from S3 storage using a
shellcommandProcess the downloaded GPS data by running custom Python scripts
Upload the processed GPS
.parquetfiles to the “Intermediate” bucket using ashellcommandRecompress the daily archive data and upload it to the “Raw” bucket using a
shellcommand
Within step 3, a workflow Python script (workflow/scripts/GPS_workflow.py) is exectud to process the decompressed raw .json data. This script in turn calls a GPS processing script from the Smart Health process sub-module. This Python script iterates over each individual file and processes it. This process involves:
Loading the data
Validating the data using
pydanticConverting to a
pandasdataframeAdding geometry information to facilitate distance calculations
Calculation of distance and speed measures between data points
Data cleaning of unfeasible distances or speeds. These filters can be configured.
Joining the reference LSOAs to the GPS locations
Removing columns that should not be included in the output tables (e.g. non-pseudonymised GPS data)
The LSOA is converted to
NULLat invalid time points. This is specified as after a large time jump (defaulting to 5 minutes). This prevents the inclusion of these LSOA values in aggregate calculations.Aggregation of the data at the configured time resolution (default 5 minutes) and calculation of the specified metrics
Finally, columns are renamed to match expected outputs
All of the processed data per day is then saved into one storage-efficient binary format (as Apache Parquet files). The pipeline logs the number of files successfully processed (and whether this differs from the total number of available files).
In essence, the Snakefile takes as input the GPS data .json files from the compressed daily archive, for the configured day deltas, and returns as output one .parquet file per day for each of the pseudonymised LSOA and derived mobility data.
Device Processing#
The Device Snakefile runs the following steps:
Download and extract Device data from the compressed daily archive in the “Ingress” bucket using a
shellcommandProcess the downloaded Device Daily Summary data by running custom Python scripts
Process the downloaded Device Intraday Activities data by running custom Python scripts
Combine the Daily Summary data with derived daily summary metrics from the Intraday Activities files by running custom Python scripts
Upload the processed Device
.parquetfiles to the “Intermediate” bucket using ashellcommandRecompress the daily archive data and upload it to the “Raw” bucket using a
shellcommand
Within steps 2,3, and 4, a workflow Python script (workflow/scripts/XX_workflow.py) is executed to process the decompressed raw .json data. Here, XX is either fitbit_intraday_, fitbit_summary, or combine_summaries. The fitbit_intraday_ and fitbit_summary scripts in turn call processing scripts from the Smart Health process sub-module. These Python script iterate over each individual file and processes it by:
Loading the data
Validating the data using
pydanticCreating a
pandasdataframe
For the intraday data processing, an additional step exists to: 4. Aggregate the data into both:
Intraday measures (default 5 minute intervals)
A daily summary
All of the processed data per day is then saved into one storage-efficient binary format (as Apache Parquet files). At this stage, the Daily Summary data is an ‘Interim’ file. The pipeline logs the number of files successfully processed (and whether this differs from the total number of available files).
The combine_summaries workflow then joins the daily aggregated metrics from the raw intraday data into the Interim Daily Summary dataframe, before saving the combined output into a final Daily Summary .parquet file.
The Snakefile takes as input the Device data Daily Summary and Intraday Activities .json files from the compressed daily archive, for the configured day deltas, and returns as output one .parquet file per day for the Daily Summary data and one file for each of the Intraday Activities.
Stage 2: Intermediate to Clean#
Click here to see the Stage 2 pipeline diagram
Stage 2 of the Smart Health pipeline#
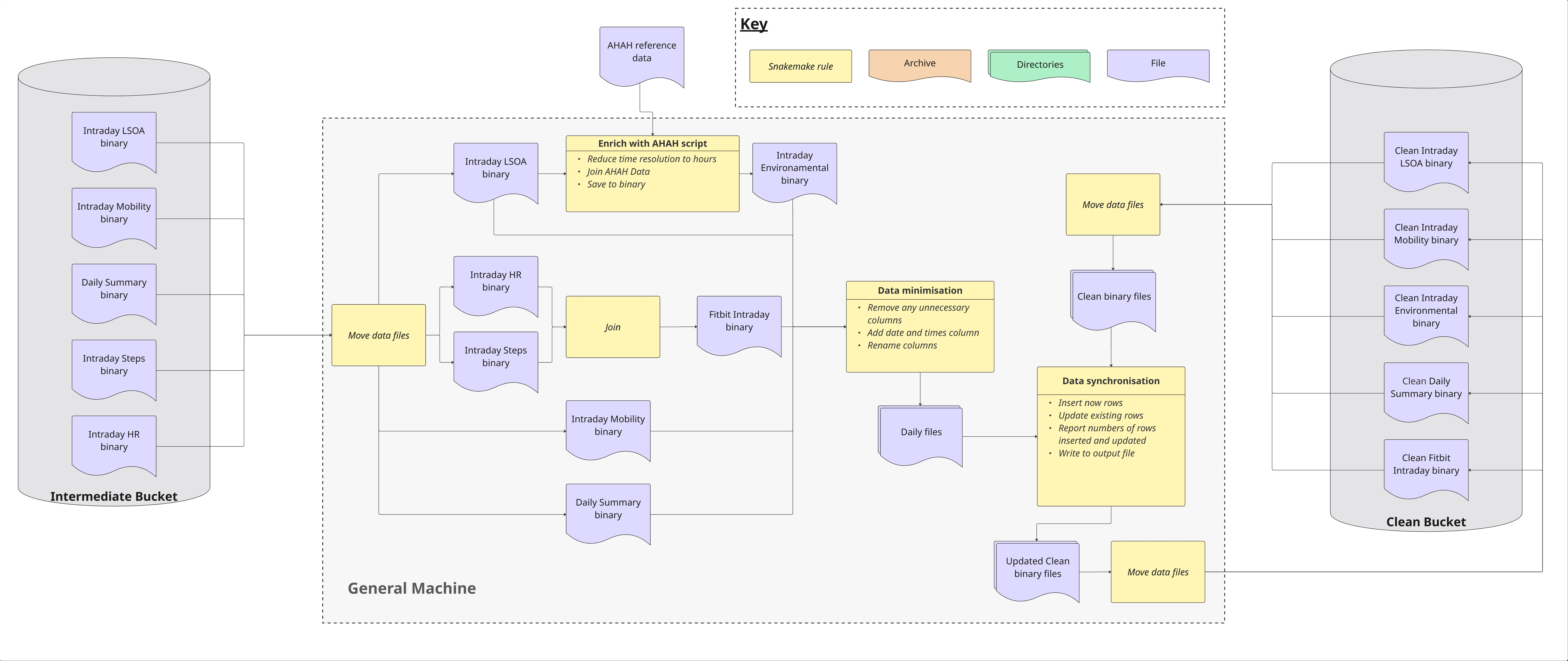
A diagram outlining the Snakemake rules, input and output files, and data flow of Stage 2 of the Smart Health pipeline#
As it is pseudonymised data, the LSOA data can now be processed on the same machine as other data streams. Therefore, one Snakefile co-ordinates Stage 2 of the Smart Health pipeline.
Data Enrichment#
Data Synchronisation#
Customising the Pipeline#
Please see Getting Started for a quickstart guide to running the Smart Health pipeline.
Snakemake configuration#
Snakemake can be configured using the keys available in workflow/config/config.yaml.
Snakemake can also be configured further by using command line arguments. For a full list of available options, see the Snakemake documentation
Day Deltas#
Two settings are available:
GPS_DAY_DELTASDEVICE_DAY_DELTASBoth keys expect a list of integers which indicate the days prior to when the archive was recieved that should be processed. Note that this should always be a value greater than 1. Values less than 1, or repeated values, will be ignored.
Output paths#
LOCAL_OUTPUT_PATH defines the local machine path for storing intermediate files.
LOCAL_LOG_PATH defines the local machine path for storing log files. A subdirectory called logs/ will be created unter this directory.
Database configuration#
The Smart Health database can be configured using src/bhf_smarthealth/conf.py.
Logging level#
The logging configuration can be set using DEFAULT_LOG_LEVEL, set to INFO by default.
See [logging levels] (https://docs.python.org/3/library/logging.html#logging-levels) for available levels.
Intraday Aggregation Period#
The time resolution that intraday data (activities, LSOA, and mobility) is aggregated to is specified by DEFAULT_TIME_PERIOD, which defaults to 5 minutes.
Data Output Settings#
A number of settings exist to customise the data ouputs of the Smart Health pipeline. This includes the aggregation measures (e.g. mean, median, min, max etc.), which columns are aggregated (this can be more than one aggregate measure), and the name of the columns in the output files.
GPS data specific settings#
DEFAULT_GPS_FILE_NAME: specifies the file name of the raw .json GPS data file to expect. Defaults to gps_data.
DEFAULT_GPS_CRS: The co-ordinate reference system (CRS) to map GPS latitude and longitude values to. Defaults to “EPSG:4326”.
DEFAULT_PLANAR_CRS: The planar CRS to map GPS datapoints to. Necessary for distance calculations. This defaults to the British National Grid.
DEFAULT_DECIMAL_PLACES: The default decimal places to round to. Defaults to 3.
DEFAULT_MAX_SPEED_MS: The maximum allowed speed value, in metres per second. Defaults to 36 m/s, or ~130 kph. Values above this value are filtered from the data set.
DEFAULT_MAX_TIME_JUMP_MINS: The maximum allowed time jump, in minutes, between data points. Default to 5 minutes. If a time jump between data points is greater than this value, the distance and speed of travel between the adjacent points is omitted from the dataset. The LSOA values corresponding to these data points will also not be included in aggregate calculations.
DEFAULT_REMOVE_GPS_COLUMNS: The default columns to remove from the processed GPS pandas dataframe. Defaults to ["lat", "long", "geometry", "index_right"]. The first three must be removed to remove the highly-sensitive GPS data, whilst the index_right is a superfluous field generated during data joining.
DEFAULT_GPS_AGG_DICT: The Python dictionary of columns to aggregate, and the calculations to perform for each column. Defaults to
{
"distance": ["size", "sum"],
"speed": ["mean"],
"LSOA21NM": ["last"],
"LSOA21CD": ["last", "nunique"],
}
DEFAULT_RENAME_GPS_COLUMNS: Defines the column names in the output tables. Defaults to
[
"mobility_n_samples",
"dist_travelled",
"mean_speed",
"latest_LSOA21NM",
"latest_LSOA21CD",
"n_unique_LSOAs",
]
DEFAULT_GPS_OUTPUT_COLUMNS: Defines the output files and their columns. Defaults to
{
"LSOA_out": ["ID", "latest_LSOA21CD", "latest_LSOA21NM"],
"mobility_out": ["ID", "mobility_n_samples", "dist_travelled", "mean_speed", "n_unique_LSOAs"],
}
Device data specific settings#
DEFAULT_FB_INTRADAY_AGG_DICT: The Python dictionary of columns to aggregate, and the calculations to perform for each column. Defaults to
{
"heart": {"value": ["mean", "min", "max", "size"]},
"steps": {"value": ["sum", "size"]},
}
DEFAULT_FB_INTRADAY_AGG_SETTINGS: The time periods to aggregate the Fitbit intraday activities to. Facilitates the generation of additional Daily Summary data derived from the intraday raw data. Defaults to:
{
"intraday": {"time_period": DEFAULT_TIME_PERIOD},
"summary": {"time_period": "1d", "label": "left"},
}
Environmental data specific settings#
DEFAULT_AHAH_TIME_PERIOD: The time period to aggregate LSOA data into to join with environmental measures. Defaults to 1 hour.
DEFAULT_AHAH_AGG_DICT: The Python dictionary of columns to aggregate, and the calculations to perform for each column. Defaults to
{
"LSOA_n_samples": ["sum"],
"latest_LSOA21CD": [aggregators.mode_with_later_tiebreak],
"latest_LSOA21NM": [aggregators.mode_with_later_tiebreak],
}
DEFAULT_RENAME_AHAH_COLUMNS: Defines the column names in the output tables.
[
"env_n_samples",
"modal_LSOA21CD",
"modal_LSOA21NM",
]
DEFAULT_AHAH_MERGE_COL: The column used to merge reference AHAH data with the aggregated LSO data. Defaults to [“modal_LSOA21CD”].
DEFAULT_REMOVE_AHAH_COLUMNS: The default columns to remove from the Environmental data table. Defaults to ["LSOA21CD", "modal_LSOA21CD", "modal_LSOA21NM"].
DEFAULT_AHAH_FIELDS: Defines the AHAH data fields available in the output Environmental data table.
